
Shubhra Ghosh Dastidar
Professor
Shubhra Ghosh Dastidar
Professor, Biological Sciences
PhD: University of Calcutta, 2006
Previous appointments:
2005-2006: Postdoctoral Researcher at University of California (Davis), CA, USA, in the group of Yong Duan.
2007: Postdoctoral Researcher University of Texas Medical Branch at Galveston, TX, USA, in the group of Catherine Schein and Werner Braun
2007-2010: Postdoctoral Researcher at Bioinformatics Institute, A*STAR, Singapore, in the group of Chandra Verma
Research interests:
Broader areas of interest: Computational Chemistry and Biology, Bioinformatics, Drug Design, Data Science, Machine Learning and AI, Structure and Dynamics of Biomolecules, Molecular Modelling and Simulations
Biomolecules and their assemblies
are complex chemical systems. Understanding their mechanisms of functions require
various methods of investigations, in truly interdisciplinary mode, covering theoretical,
experimental, computational methods and even data analytics. In our group we address
those questions on the mechanism of functions Biomolecular systems, which would
require insights not only from the molecular structures but also would need the
information on the dynamics of the constituent atoms; in fact in real systems the
atoms and molecules in a cell are always jiggling, dancing and bumping into
each other at any given temperature. For example, one may figure out a model structure
of a protein from the experimental measurement or may obtain from from a structure
database, but that may not be sufficient to guess the mechanical flexibility of
the system which could have relation with the function. The dynamics could be
simulated in computer using fundamental principles of Chemistry, Biochemistry, Physics
and the obtained datasets are analyzed using statistical methods to interpret
the observations in Life sciences. The computations are done on large scale
parallel computing platforms, often using GP-GPU based machines.
Our systems of specific interests are proteins, lipids, nucleic acids, small molecule (drugs). These exercises only tells us how do the biomolecules function but also lead to understand the kind of errors at the molecular level that can result functional defect and diseases. Once this is understood, it provides a direction for designing a lead compound (drug molecule) in a rational manner. Some of our recent interests include (i) mechanism of conformational modulations on αβ-Tubulin dimer and their control over cell divisions, (ii) structural dynamics of the Bcl2 family of proteins and their role in apoptosis and cancer therapy, (iii) Kinase allosteric and their inhibition using drug molecules, (iv) ligand design and various others.
Primarily we use standard molecular dynamics methods to simulate the all-atom dynamics of the systems and also use more advanced methods of accelerated molecular dynamics to compute the free energy landscapes of the biomolecular functions. We are also developing machine learning based protocols to analyze conformational ensembles.
Here is a list of selected publications:
J Chem Inf Model 2025
Biochemistry, 2024, 63, 11, 1474–1492
Comput Biol Chem. 2024;108:108004
Proteins. 2023, doi: 10.1002/prot.26634
J Chem Inf Model. 2023, 63, 224-239.
Comput Biol Chem 2022; 96, 107617.
Biochemistry 2020, 59, 45, 4353
J. Chem. Inf. Model. 2019, 59, 5, 2274
J. Chem. Inf. Model. 2018, 58, 2, 370
J. Phys. Chem. B 2017, 121, 1, 118
Biochemistry 2016, 55, 2, 335
Phys Chem Chem Phys 2016, 18, 24095
Biochemistry 2012, 51, 36, 7138
Click here for the complete list
(Movies, with specific scientific insights, are available as the supporting information of these articles, which are usually available free of charges from the journal websites)
Group Members (current): Dr. Debarati Paul (Project Scientist), Megha Ghosh (Senior Research Fellow), Avibrata Dutta (Junior Research Fellow), Payel Bhanja (Junior Research Fellow), Mrs. Munmun Dey (Scientific Administrative Assistant, project funded)
Ph.D. positions: Our group is looking for at least 2-3 motivated students. Candidates with Masters in Chemistry/ Biophysics/ Biochemistry/ Physics/ Biotechnology/ Statistics/ Computer Applications, etc. and with NET-JRF or equivalent qualification for fellowship may send their CV by email to sgd[at]jcbose.ac.in.
Postdoc/RA positions: Currently we are looking for candidates with knowledge on Machine Learning based data analytics or at least the candidate who have interest to learn these methods to make applications. Anyone willing to apply for N-PDF (SERB)/ DBT-RA or equivalent, may contact by email.
Contact:
| Address: |
Biological Sciences Unified Academic Campus Bose Institute EN-80, Sector V Bidhan Nagar Kolkata - 700 091, India |
| E-Mail: | sgd[at]jcbose.ac.in |
| Phone: | +91-33-25693218 |
Research:
Our recent work, a trilogy on TAK1 kinase, is accessible though the following links:
1. https://doi.org/10.1021/acs.jcim.5c00529
2. https://doi.org/10.1021/acs.biochem.3c00643
3. https://doi.org/10.1021/acs.jcim.2c00778
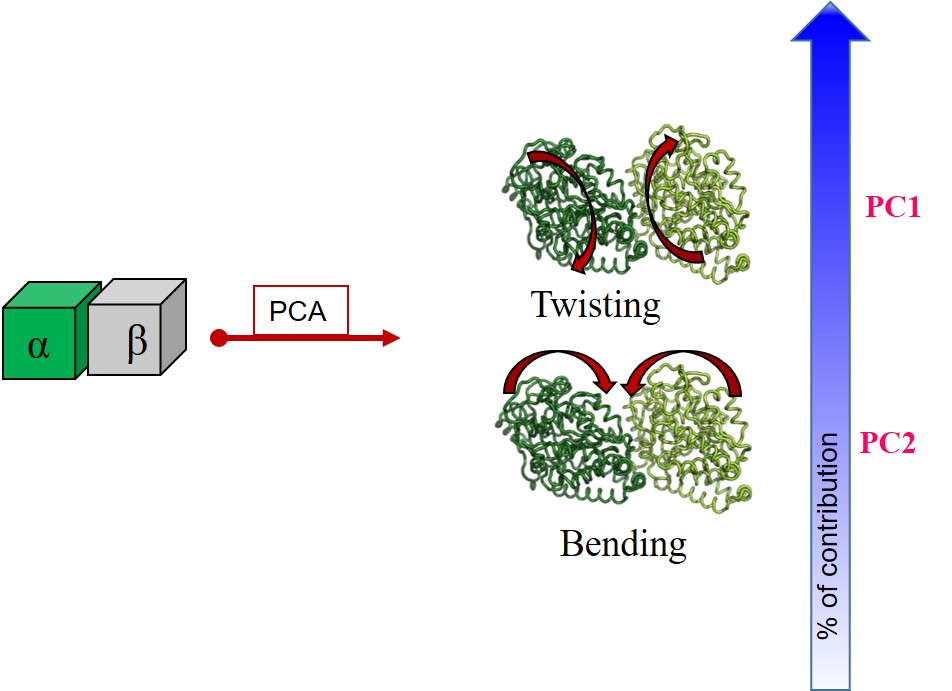
The mechanisms of ligand induced alterations in the capacity of the α,β-Tubulin dimer to form and/or not to form microtubules and spindles were revealed to be a consequence swapping between principle modes of internal motions. The details would be available in the article entitled "Ligand Binding Swaps between Soft Internal Modes of α,β-Tubulin and Alters Its Accessible Conformational Space | The Journal of Physical Chemistry B (acs.org)", J. Phys. Chem. B 2017, 121, 1, 118–128
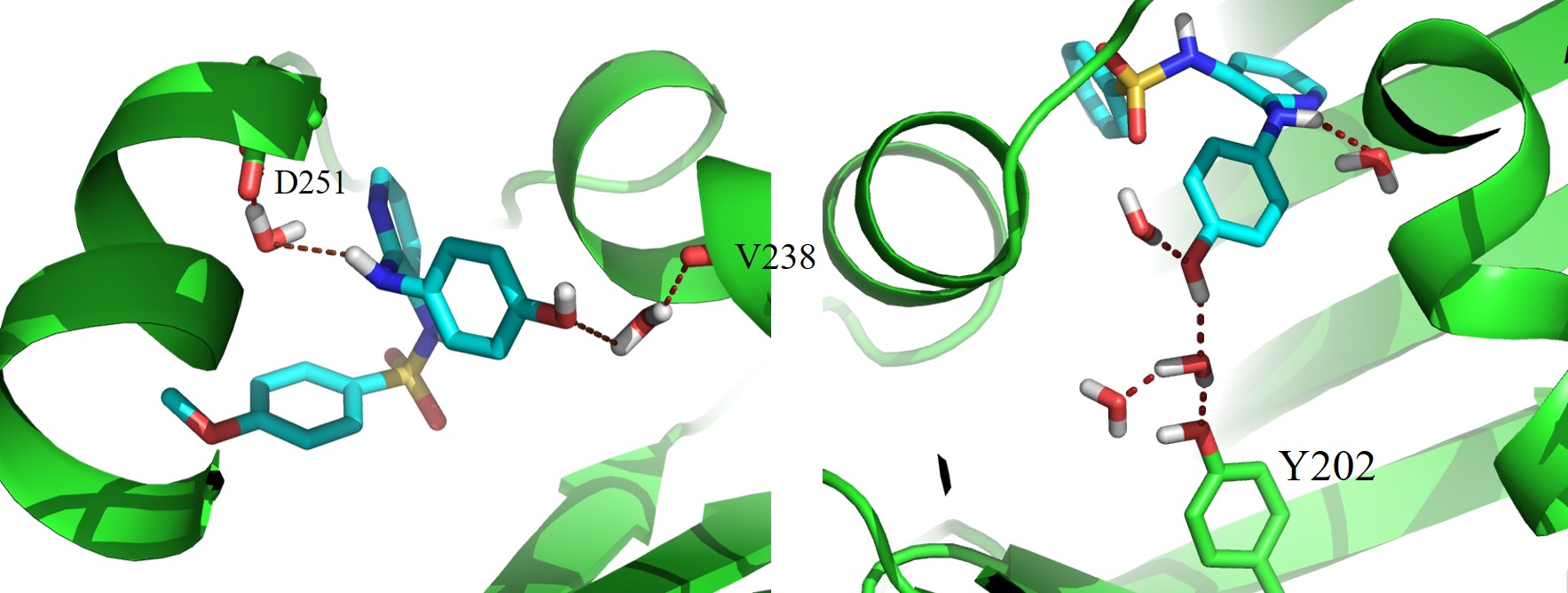
Micro-clusters of the water molecules are crucial to determine the protein-ligand binding affinities. Using α,β-Tubulin dimer and its complex with E7010 it has been shown how these water need to be included in the computation of the binding free energies. Read this article for further details: Conformational States of E7010 Is Complemented by Microclusters of Water Inside the α,β-Tubulin Core | Journal of Chemical Information and Modeling (acs.org), J. Chem. Inf. Model. 2019, 59, 5, 2274–2286
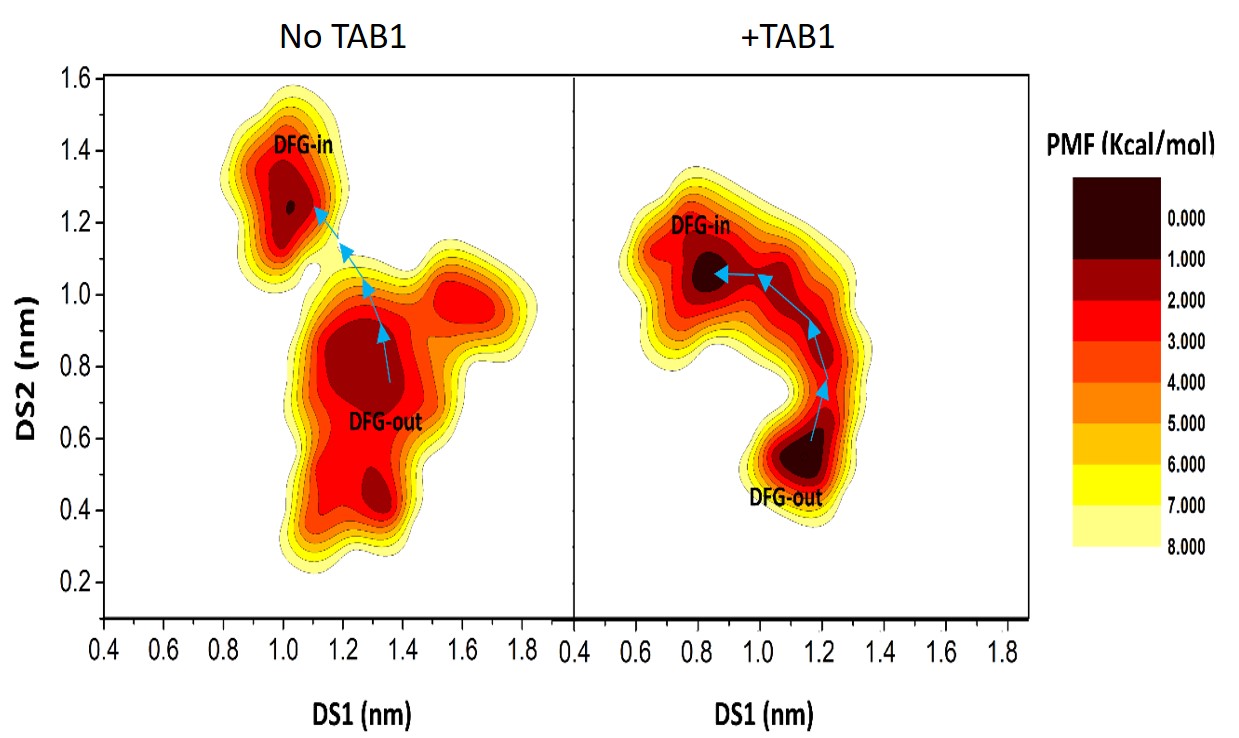
2D PMF landscape of TAK1 kinase activation, in absence (left) and presence (right) of the activator (TAB1). The work involved 5 microsec standard MD, and also TMD and GAMD, whose details would be available in the recently published article entitled "Allosteric Boost by TAB1 on the TAK1 Kinase Favorably Sculpts the Thermodynamic Landscape of Activation | Journal of Chemical Information and Modeling (acs.org)", J. Chem. Inf. Model. 2023, 63, 1, 224–239
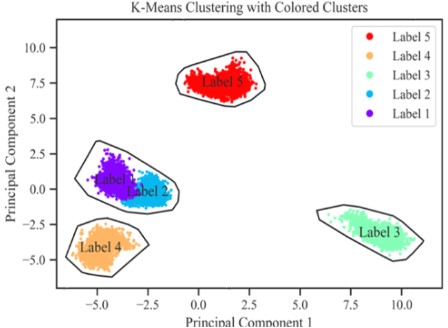
A
machine learning based classification of the conformational states of
α,β-Tubulin dimer binding 5 different ligands with different capacities to
inhibit microtubule formation. The classification enables the discrimination
between ligands in terms of their mechanism which otherwise remained difficult
to point out. Further details would be available here: "Molecular Dynamics and Machine Learning reveal distinguishing mechanisms of Competitive Ligands to perturb α,β-Tubulin - ScienceDirect", Computational Biology and Chemistry, 2024, 108, 108004
Publications:
Click here for the complete list
Publications: 2011 - 2021
1. Ray Chaudhuri N, Ghosh Dastidar S, Biochemistry, Accepted (May 2024), Adaptive Workflows of Machine Learning Illuminate the Sequential Operation Mechanism of TAK1's Allosteric Network
2. Basak P, Ray Chaudhuri N, Basu D, Ganguly D, Ghosh Dastidar S, J Biomol Struc Dyn, Accepted 2024, Molecular Origin of the Differential Stabilities of the Protofilaments in Different Polymorphs: Molecular Dynamics Simulation and Deep Learning
3. Basu D, Dastidar SG. Comput Biol Chem. 2024 Feb;108:108004. Molecular Dynamics and Machine Learning reveal distinguishing mechanisms of Competitive Ligands to perturb α,β-Tubulin.
4. Dasgupta S, Dev A, Chongdar N, Basak P, Dastidar SG, Basu G. Proteins. 2023 Nov 12. doi: 10.1002/prot.26634.Signatures of tRNAGlx -specificity in proteobacterial glutamyl-tRNA synthetases.
5. Seal S, Chakraborty T, Polley S, Paul D, Banerjee N, Sinha D, Dutta A, Chatterjee S, Sau K, Ghosh Dastidar S, Sau S. J Biomol Struct Dyn. 2023 Oct 30:1-14. Modeling and monitoring the effects of three partly conserved Ile residues in the dimerization domain of a Mip-like virulence factor from Escherichia coli.
6. Kar S, Maji N, Sen K, Roy S, Maity A, Ghosh Dastidar S, Nath S, Basu G, Basu M. Biosci Rep. 2023 Aug 31;43(8):BSR20230677.Reprogramming of glucose metabolism via PFKFB4 is critical in FGF16-driven invasion of breast cancer cells.
7. Roy A, Basu D, Bose D, Dutta A, Dastidar SG, Chatterjee S., Int J Biol Macromol. 2023; 231:123263. Identification and characterization of a flexile G-quadruplex in the distal promoter region of stemness gene REX1.
8. Ray Chaudhuri N, Ghosh Dastidar S., J Chem Inf Model 2023;63(1):224-239. doi: 10.1021/acs.jcim.2c00778. Allosteric Boost by TAB1 on the TAK1 Kinase Favorably Sculpts the Thermodynamic Landscape of Activation.
9. Banerjee N, Chatterjee O, Roychowdhury T, Basu D, Dutta A, Chowdhury M, Dastidar SG, Chatterjee S., Biochim Biophys Acta Gen Subj. 2023;1867(2):130267. Sequence driven interaction of amino acids in de-novo designed peptides determines c-Myc G-quadruplex unfolding inducing apoptosis in cancer cells.
10. Mondal A, Paul D, Dastidar SG, Saha T, Goswami AM., Sci Rep. 2022; 12(1):14934.In silico analyses of Wnt1 nsSNPs reveal structurally destabilizing variants, altered interactions with Frizzled receptors and its deregulation in tumorigenesis.
11. Paul D, Basak P, Ghosh Dastidar S., Comput Biol Chem. 2022; 100:107736. Remote communication between unstructured and structured regions of Bcl-2 tunes its ligand binding capacity: Mechanistic insights.
12. Ghosh A, Kar PK, Gautam A, Gupta R, Singh R, Chakravarti R, Ravichandiran V, Ghosh Dastidar S, Ghosh D, Roy S., RSC Med Chem. 2022; 13(6):647-675.An insight into SARS-CoV-2 structure, pathogenesis, target hunting for drug development and vaccine initiatives.
13. Basu D, Majumdar S, Mandal N, Dastidar SG., Comput Biol Chem. 2022; 96:107617. Mechanisms of influence of the microtubule over-stabilizing ligands on the structure and intrinsic dynamics of α,β-Tubulin.
14. De A, Maity A, Mazumder M, Mondal B, Mukherjee A, Ghosh S, Ray P, Polley S, Ghosh Dastidar S, Basu D., Plant Sci. 2021, 309:110953, Overexpression of LYK4, a lysin motif receptor with non-functional kinase domain, enhances tolerance to Alternaria brassicicola and increases trichome density in Brassica juncea.
15. Paul D, Basu D, Ghosh Dastidar S., J Mol Model. 2021;27(5):128. Multi-conformation representation of Mpro identifies promising candidates for drug repurposing against COVID-19.
16. Sinha S, Ghosh Dastidar S., Biochemistry 2020; 59(45):4353-4366. Shifting Polar Residues Across Primary Sequence Frames of Transmembrane Domains Calibrates Membrane Permeation Thermodynamics.
17. Bhattacharyya R, Dhar J, Ghosh Dastidar S, Chakrabarti P, Weiss MS., IUCrJ. 2020;7(Pt 5):825-834.’ The susceptibility of disulfide bonds towards radiation damage may be explained by S⋯O interactions.
18. Shohan MUS, Sinha S, Nabila FH, Ghosh Dastidar S, Seraj ZI. Front Plant Sci. 2019 Nov 4;10:1420. HKT1;5 Transporter Gene Expression and Association of Amino Acid Substitutions With Salt Tolerance Across Rice Genotypes.
19. Meher G, Sinha S, Pattnaik GP, Ghosh Dastidar S, Chakraborty H. J Phys Chem B. 2019 Aug 22;123(33):7113-7122. Cholesterol Modulates Membrane Properties and the Interaction of gp41 Fusion Peptide To Promote Membrane Fusion.
20. Maity A, Sinha S, Ghosh Dastidar S., Chem Phys Lipids. 2019 ;218:112-124., Dissecting the thermodynamic contributions of the charged residues in the membrane anchoring of Bcl-xl C-terminal domain.
21. Majumdar S, Basu D, Ghosh Dastidar S., J Chem Inf Model. 2019;59(5):2274-2286. , Conformational States of E7010 Is Complemented by Microclusters of Water Inside the α,β-Tubulin Core.
22. Chakraborty J, Priya P, Ghosh Dastidar S, Das S., Plant Sci. 2018;276:111-133., Physical interaction between nuclear accumulated CC-NB-ARC-LRR protein and WRKY64 promotes EDS1 dependent Fusarium wilt resistance in chickpea.
23. Maity A, Majumdar S, Ghosh Dastidar S., Comput Biol Chem. 2018 Dec;77:17-27. , Flexibility enables to discriminate between ligands: Lessons from structural ensembles of Bcl-xl and Mcl-1.
24. Sinha S, Maity A, Ghosh Dastidar S., J Chem Inf Model. 2018 Feb 26;58(2):370-382. , BIM Binding Remotely Regulates BAX Activation: Insights from the Free Energy Landscapes.,
25. Basak P, Maitra-Majee S, Das JK, Mukherjee A, Ghosh Dastidar S, Pal Choudhury P, Lahiri Majumder A., PLoS One. 2017 Sep 26;12(9):e0185351. , An evolutionary analysis identifies a conserved pentapeptide stretch containing the two essential lysine residues for rice L-myo-inositol 1-phosphate synthase catalytic activity.
26. Priya P, Maity A, Ghosh Dastidar S., Proteins Struc Func Bioinf 2017 ;85(8):1567-1579. , The long unstructured region of Bcl-xl modulates its structural dynamics.
27. Majumdar S, Ghosh Dastidar S., J Phys Chem B. 2017;121(1):118-128., Ligand Binding Swaps between Soft Internal Modes of α,β-Tubulin and Alters Its Accessible Conformational Space.
28. Maity A, Sinha S, Ganguly D, Ghosh Dastidar S., Phys Chem Chem Phys. 2016 ;18(34):24095-105. C-terminal tail insertion of Bcl-xL in membrane occurs via partial unfolding and refolding cycle associating microsolvation.
29. Majumdar S, Maiti S, Ghosh Dastidar S, Biochemistry 2016; 55, 335-47. Dynamic and Static Water Molecules Complement the TN16 Conformational Heterogeneity inside the Tubulin Cavity.
30. Sinha A, Ray A, Ganguly S, Ghosh Dastidar S, Sarkar S., Biol Direct. 2015;10(1):56. Variation in the ribosome interacting loop of the Sec61α from Giardia lamblia.
31. Bhar K, Maity A, Ghosh A, Das T, Ghosh Dastidar S, Siddhanta A., Protein J. 2015 Apr;34(2):158-67., Phosphorylation of Leghemoglobin at S45 is Most Effective to Disrupt the Molecular Environment of Its Oxygen Binding Pocket
32. Priya P, Maity A, Majumdar S, Ghosh Dastidar S., J Mol Graph Model. 2015;59:1-13. Interactions between Bcl-xl and its inhibitors: Insights into ligand design from molecular dynamics simulation.
33. Maity A, Majumdar S, Priya P, De P, Saha S, Ghosh Dastidar S, J Biomol Struct Dyn. 2015;33(2):298-321 Adaptability in protein structures: Structural dynamics and implications in ligand design (Review)
34. Maity A, Yadav S, Verma CS, Ghosh Dastidar S, PLoS One (2013) 8, e76837. Dynamics of Bcl-xl in Water and Membrane: Molecular Simulations
35. Sengupta A, Sarkar A, Priya P, Dastidar SG, Das S, PLoS One 2013, 8, e78249. New Insight to Structure-Function Relationship of GalNAc Mediated Primary Interaction between Insecticidal Cry1Ac Toxin and HaALP Receptor of Helicoverpa armigeraí.
36. Chakraborti S, Chakravarty D, Gupta S, Chatterji BP, Dhar G, Poddar A, Panda D, Chakrabarti P, Ghosh Dastidar S* and Bhattacharyya B*, Biochemistry 2012, 51,7 138, Discrimination of Ligands with Different Flexibilities Resulting from the Plasticity of the Binding Site in Tubulin
37. Dastidar SG, Lane DP, Verma CS, Cell Cycle 2012, 11: 2239-47, Why is F19Ap53 unable to bind MDM2? Simulations suggest crack propagation modulates binding
38. C. J. Brown£, S. G. Dastidar£, S. T. Quah, A. Lim, B. Chia, C. S. Verma. PLoS One 2011, 6, e24122, C-Terminal Substitution of MDM2 Interacting Peptides Modulates Binding Affinity by Distinctive Mechanisms
39. G. Fuentes, S. G. Dastidar, A. Madhumalar, C. S. Verma, Drug Dev. Res. 2011, 72, 26, Role of protein flexibility in the Discovery of New Drugs (Review Article)
40. S. G. Dastidar, D. Raghunathan, J. Nicholson, T. R. Hupp, D. P. Lane, C. S. Verma, Cell Cycle 2011, 10, 82, Chemical States of the N-terminal “lid” of MDM2 regulate p53 binding
Click here for the complete list
Recognition:
- Associate Editor, Frontiers in Molecular Biosciences,
Teaching:
Students:
| Image | Name | Designation | Department | Campus | Contact number | |
|---|---|---|---|---|---|---|
 |
Avibrata Dutta | Junior Research Fellow | Biological Sciences | Unified | avibratadutta007@gmail.com | |
 |
Megha Ghosh | Junior Research Fellow | Bioinformatics Centre | Unified | ghoshmegha39@gmail.com | |
 |
Payel Bhanja | Junior Research Fellow | Biological Sciences | Unified | payelbhanja34@gmail.com |
Former:

Ex students who have already been awarded with the doctoral degree:
1. Nibedita Ray Chaudhuri, Ph.D. (2025), Research Scientist, Quantiphi, Bangalore
2. Debarati Paul, Ph.D. (2024), Project Scientist (DBT), Bose Institute, Kolkata
3. Debadrita Basu, Ph.D. (2024), Research Scientist, Sravathi AI technology Pvt Ltd., Bangalore
4. Souvik Sinha, Ph.D. (2021), Postdoctoral Fellow, University of California at Irvine, USA
5. Sarmistha Majumdar, Ph.D. (2019), Scientist-I at Schrödinger, Inc. Hyderabad
6. Prerna Priya, Ph.D. (2019), Guest Lecturer at Department of Botany, Purnea University, India
7. Atanu Maity, Ph.D. (2018), Project Scientist (DBT), IIT Kharagpur, India

Trainee/Interns, etc.
To be updated .......
Group News:

Interviews and popular science talk shows in TV channels:
1. https://www.youtube.com/live/3qV1T6D9EJI?si=3XFasYr2fqlz-wcm
2. https://www.youtube.com/live/nd2SxFfqCBY?si=skDr3V5Iirstd4H6
Updates:
2025
Nibedita Ray Chaudhuri has been awarded with the PhD degree
Premananda Basak has submitted thesis for PhD degree
2024
Dr. Debarati Paul has joined as a Project Scientist
Mrs. Munmun Dey has joined as a Scientific Administrative Assistant (DBT project funded)